X-ray analysis of yeast lipoamide
dehydrogenase complexed with NAD+
Wataru Adachi,a Kaoru Suzuki,b Masaru
Tsunoda,c Takeshi Sekiguchi,b Lester J. Reedd and Akio
Takénakaa
aGraduate
School of Bioscience and Biotechnology, Tokyo Institute of Technology, Nagatsuta, Midori-ku, Yokohama 226-8501, Japan;
bDepartment of Environmental Science,
College of Science and Engineering, Iwaki-Meisei University, Chuodai-iino,
Iwaki 970-8551, Japan; cSchool of
Pharmaceutical Sciences, Showa University, Hatanodai, Shinagawa, Tokyo
142-8555, Japan; dDepartment of Chemistry
and Biochemistry, The University of Texas at Austin, Austin, TX78712, USA (wadachi@bio.titech.ac.jp).
Lipoamide
dehydrogenase (E3) is one of the components of 2-oxoacid
dehydrogenase complex. This enzyme catalyses the oxidization of a dihydrolipoyl
group of E2 with the help of the cofactors, NAD+ and
FAD. E3 belongs to the pyridine nucleotide-disulphide oxidoreductase family of glutathione reductase, trypanothione reductase, mercuric
ion reductase, etc. We already
reported the native E3 structure from Saccharomyces cerevisiae [1].
In this study, we co-crystallized yeast E3 with NAD+
to elucidate the mechanism of substrate binding.
Diffraction data were collected at
100K and processed at 2.2 Ā resolution. The space group and cell parameters are
P212121, a=66.6,
b=96.4, and c=160.0Ā,
respectively. Initial phases were
derived by molecular replacement using the native structure. The atomic parameters were refined (the
final R=20.3% and Rfree=24.6%).
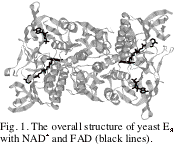
The overall structure of yeast E3-NAD+
complex is shown in Fig. 1. When
the structure is superimposed on the native one, the
overall root-mean-square deviation is 0.62 Ā, suggesting no significant difference on NAD+
binding. The final electron
density map clearly indicates that the adenosine moiety and the pyrophosphate
group of NAD+ are bound to the enzyme, but the remaining
nicotinamide moiety is disordered (see Fig. 2).
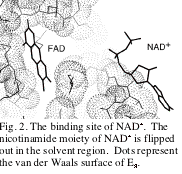
 It is
interesting to note that the binding mode of the nicotinamide moiety is
different from those of the related enzymes in the same family. Contrary in the structures of those
enzymes complexed with NAD(P)H (reduced form), the nicotinamide moiety is stacked
on the isoalloxazine ring of FAD for electron transfer, and the side chain of a
Tyr residue near the binding pocket is flip out. In the present enzyme, however, the nicotinamide does not
enter the binding site, despite that the Tyr residue is replaced with Ile. It is concluded that the exact
positioning of NAD+ on FAD depends on the redox state of the
cofactor rather than on the existence of the Tyr side chain.
It is
interesting to note that the binding mode of the nicotinamide moiety is
different from those of the related enzymes in the same family. Contrary in the structures of those
enzymes complexed with NAD(P)H (reduced form), the nicotinamide moiety is stacked
on the isoalloxazine ring of FAD for electron transfer, and the side chain of a
Tyr residue near the binding pocket is flip out. In the present enzyme, however, the nicotinamide does not
enter the binding site, despite that the Tyr residue is replaced with Ile. It is concluded that the exact
positioning of NAD+ on FAD depends on the redox state of the
cofactor rather than on the existence of the Tyr side chain.
References
1 (1998) J. Biochem. 123, 668-674